Proteins for the MCAT: Everything You Need to Know
/Learn key MCAT concepts about proteins, the structure and classification of amino acids, plus practice questions and answers
(Note: This guide is part of our MCAT Biochemistry series.)
Table of Contents
Part 1: Introduction to proteins
Part 2: Amino acids
a) Structure of amino acids
b) Classifying amino acids
Part 3: Peptide bonds
a) Formation
b) Hydrolysis
Part 4: Protein structure
a) Primary, secondary, tertiary, and quaternary structures
b) Specialized amino acids
c) Stability and interactions
d) Protein folding
Part 5: High-yield terms
Part 6: Passage-based questions and answers
Part 7: Standalone questions and answers
----
Part 1: Introduction to proteins
Proteins are an incredibly high-yield concept on the MCAT, but like a lot of biochemistry topics, they aren’t easily mastered without a great deal of practice. These topics are especially intimidating because there is virtually no limit to what you can learn about proteins, amino acids, and everything else.
This guide will serve as an introduction to amino acids, protein structure, and protein interactions. While it will not be a comprehensive handbook to everything about proteins, it will be a good place to start studying these basic principles of biochemistry. Be sure to refer to our other biochemistry guides for further information on proteins, enzymes, and other biological molecules.
Throughout the guide, you will encounter several bolded terms. Their definitions are particularly important and can also be found in Part 4 of this guide. At the end of this guide, you will also find several passage-based and standalone questions to sharpen your skills.
Let’s begin!
----
Part 2: Amino acids
a) Structure of amino acids
Amino acids are the building blocks of all proteins. The structures of amino acids are an extremely high-yield topic to study.
The structure of each amino acid can be divided into three separate regions:
The amino group, or N-terminus
The carboxylic acid group, or C-terminus
A unique identifying side chain, or R-group
Figure: Each amino acid has an amino group, an R group, and a carboxylic acid group.
Recall that an amino group is a functional group composed of NH3+. It is similar to ammonium (NH4+), except that at one position, the nitrogen is attached to a carbon instead of a hydrogen (NH2C instead of NH3). Note that this results in one free electron pair on the nitrogen atom. At physiological pH (pH ~7), this free electron pair is able to accept a bond to a single hydrogen atom. This results in a positive charge on the functional group.
The acid on every amino acid is a carboxylic acid, a functional group composed of COOH. At physiological pH (pH ~7), this carboxylic acid is deprotonated, leaving a negative charge on the functional group.
Note that at physiological pH, amino acids are zwitterions; they contain both positive and negative charges on the same molecule. Most amino acids have a net charge of zero. Exceptions arise when accounting for charges on the R-group, or side chain, of the amino acid.
These R groups, or side chains, can be as simple as a single hydrogen atom or as complex as an imidazole ring. There are 20 different R groups—each of which you should commit to memory. We’ll discuss these side chains further in the next section.
The R group is connected to the central carbon, which is known as the alpha carbon. This carbon is connected to every constituent of the amino acid: the amino group (-NH3+), the carboxylic acid part (-COO-), the R group, and a hydrogen atom (H).
Note that for 19 of the 20 amino acids, the alpha carbon itself is chiral, or attached to four different constituent groups. (The exception happens to be glycine, as the R group is simply a hydrogen atom.) Chirality refers to right- or left-handedness, denoted as D- and L- molecules, respectively. The chirality of biological molecules becomes quite important, as only L-configuration (left-handed) amino acids can be used by the body. (D-amino acids are not naturally found in eukaryotic metabolic pathways.)
Daltons (Da) are the unit of mass used to describe the masses of amino acids and proteins. They are defined as equivalent to Atomic Mass Units and therefore, 1 Dalton = 1 AMU = 1 g/mol. The average mass of an amino acid is 110 Da - this is useful for estimating the mass of a protein based on the number of amino acids in its peptide chain. For example, a protein formed from 100 amino acids would have a mass of 11,000 Da or 11 kDa.
Furthermore, some questions might ask us how many grams are present for a mole of such a protein. In this case, we simply recall that each Dalton = 1 g/mol. Our 100 amino acid example has a mass of 11,000 Da, meaning each mole of protein of this protein weighs 11,000 grams! To recap, just keep in mind that each amino acid is 110 Daltons, or 110 g/mol. This should help tackle any questions related to Daltons. As a final note, it is often ok to round 110 to 100, to make calculations easier - just be sure to remember this estimation when checking for the right answer!
b) Classifying amino acids
Each amino acid has a characteristic side chain, and the properties of these side chains are essential for the function of proteins.
Figure: A table of amino acids, including three- and one-letter abbreviations, side chains (highlighted), and the pKa of any acidic or basic side chains.
Note that there are three ways to refer to an amino acid: by its full name, its three-letter abbreviation, or its one-letter abbreviation. The MCAT may test your knowledge of all three, so be sure to memorize each form.
Nonpolar side chains
There are ten nonpolar amino acids: glycine, alanine, phenylalanine, valine, leucine, isoleucine, tyrosine, tryptophan, proline, and methionine. These side chains are considered nonpolar due to their aliphatic nature; they are primarily composed of carbons and hydrogens.
Due to their nonpolar nature, these amino acids will not be attracted to water surrounding the protein. Nonpolar amino acids are often located in the core of proteins (proteins are 3-dimensional) in aqueous solution. These nonpolar amino acids are essential to forming the structure of the protein.
Polar side chains
There are five polar amino acids: serine, threonine, asparagine, cysteine, and glutamine. Due to the attraction between water and other polar molecules, these amino acids are often positioned on the outside of the protein or the protein’s active site. Polar side chains will be attracted to other polar side chains and to water molecules in the aqueous solution that surrounds the protein.
Acidic side chains
There are two acidic amino acids: aspartate and glutamate. These residues are proton donors. They will often lose a proton in solution becoming negatively charged.
While studying for the MCAT, it’s worth noting the pKa of these acidic side chains as well. You should be able to recognize if these side chains should be protonated or deprotonated in any given range of pH values. Recall that pKa is the pH at which half of the group (or molecule) of interest is deprotonated and half is protonated. Thus, at pH=3.7, roughly half of the aspartate side chains within a protein will be deprotonated, while the other half is protonated. Thus, at pH ~ 7, most aspartates are deprotonated (and negatively charged).
Basic side chains
There are three basic amino acids: lysine, arginine, and histidine. These residues are proton acceptors. Each of these basic amino acids has a nitrogen that can accept a hydrogen to become positively charged.
As with acidic amino acids, you should also be familiar with the pKa of each of these side chains and be able to recognize if they are protonated or deprotonated within a given range of pH values.
Note that although tyrosine possesses an -OH group at the end of its side chain, its hydroxyl group is rarely found deprotonated at any physiologically relevant pH. Thus, it is considered a nonpolar amino acid--rather than an acidic one.
----
Part 3: Peptide bonds
Proteins are composed of amino acids bound together through peptide bonds. The formation of the peptide bond is catalyzed by the ribosome. As the ribosome reads an mRNA strand, it translates and adds amino acids to the growing polypeptide. You can read more about protein synthesis in our guide on RNA.
a) Formation
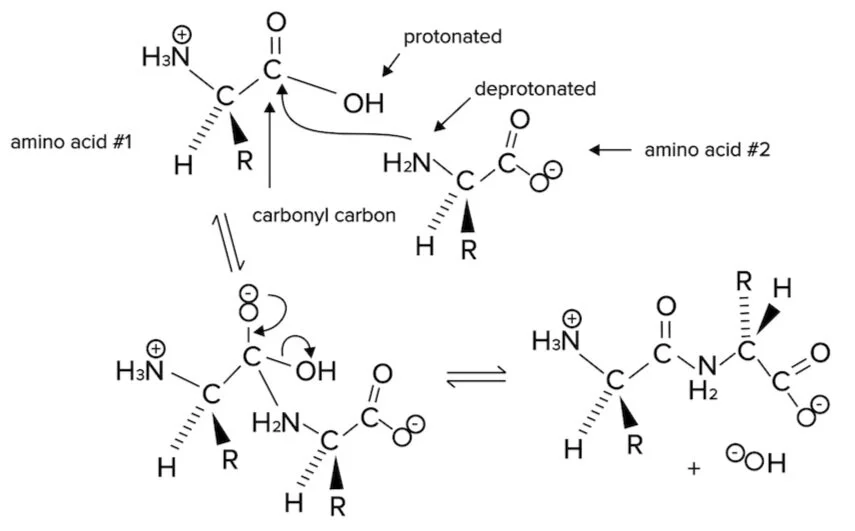
The formation of a peptide bond between two amino acids, or between an amino acid and a peptide, is an example of a nucleophilic substitution reaction (a subset of nucleophile-electrophile reaction)—a very common reaction that you should know for the MCAT.
The reaction mechanism is drawn below.
Figure: The nucleophilic substitution reaction leading to formation of a peptide bond.
Note that the carbonyl group of the carboxylic acid is an electrophile. The nucleophile is the nitrogen of the amino group. The nitrogen of the second amino acid has a free lone pair that can attack the carbonyl group, forming an N-C bond.
In the process, electrons from the second bond of the C=O are sent to the carbonyl oxygen. These electrons then reform a second bond, and the leaving group (-OH) leaves with a lone pair of electrons to form a molecule of water. This reaction can also be referred to as a dehydration reaction.
A peptide bond is formed between the nitrogen of the amino group of an amino acid and the carbon of the acid group (carboxyl) of another amino acid or growing peptide strand). This bond is an amide bond. The carbon of the amide bond is also double-bonded to an oxygen atom. Because the nitrogen of the peptide bond has a lone pair, the peptide bond has a partial double bond character.
Figure: Resonance structures of the planar amide bond.
Recall that while single bonds can freely rotate, double bonds cannot. Since the peptide bond has a partial double bond character (think of it as an average of a single and double bond), it does not rotate as much as a single bond and can be treated as a fixed bond.
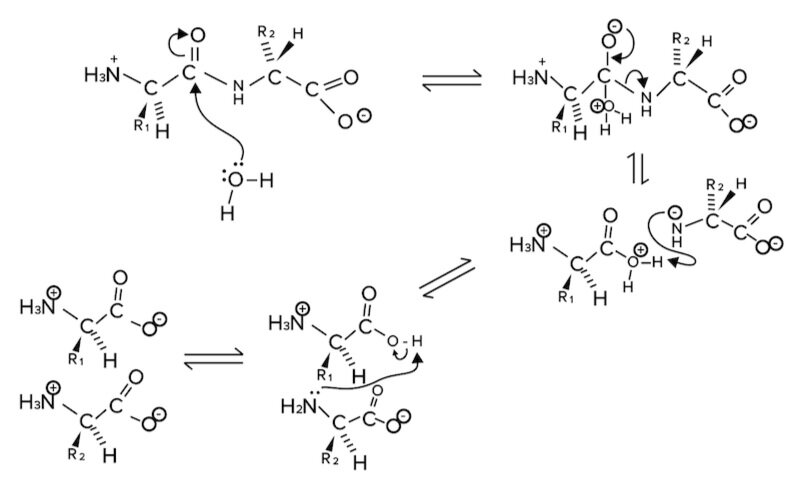
b) Hydrolysis
Let’s now discuss the reverse reaction, peptide bond hydrolysis. Hydrolysis means breaking (lysis) with water (hydro). So, peptide bonds can be broken by water molecules.
Gain instant access to the most digestible and comprehensive MCAT content resources available. 60+ guides covering every content area. Subscribe today to lock in the current investments, which will be increasing in the future for new subscribers.
Figure: Peptide bond hydrolysis.
When water (or more specifically a lone pair on the oxygen of water) attacks the carbonyl carbon, the electrons in the pi bond (double bond) move onto the oxygen atom. The nitrogen of the amide bond will leave, and the newly formed amino group will be protonated by hydrogen atoms that are in the solution.
After a hydrolysis reaction, we are left with two newly formed segments with completed amino and carboxylic acid groups on either amino acid.
----
Part 4: Protein structure
a) Primary, secondary, tertiary, and quarternary structures
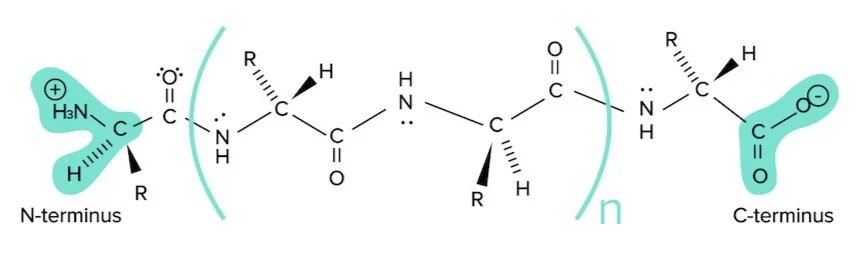
Proteins are composed of many amino acids linked together through peptide bonds. Before discussing structure, it is important to set some nomenclature. Below is a simple representation of a protein composed of n+2 amino acids.
Figure: A simple polypeptide sequence.
Notice the protein is drawn from left to right, starting with the N-terminus and ending with the C-terminus. The N-terminus refers to the side of this string with the amino group (or nitrogen) exposed. The C-terminus refers to the side of this string with the carboxylic acid group (or carbon) exposed. This is the writing convention for all protein sequences.
There are four levels of structure: primary, secondary, tertiary, and quaternary. They are all critical to the protein’s function. The first level of protein structure is its primary structure. Primary structure refers to the string of amino acids connected by peptide bonds and is defined solely by the identity of amino acids within it.
Secondary structure is formed through the hydrogen bonding interactions between atoms forming the backbone of the protein chain—rather than interactions between the side chains of each amino acid. Recall that each amino acid contains:
An N-H group, from the amide bond
A C=O bond, from the carboxylic acid
Secondary structure is composed of the hydrogen bonding interactions between the H of the N-H of one amino acid and the carbonyl oxygen (through one of its lone pairs) of another amino acid.
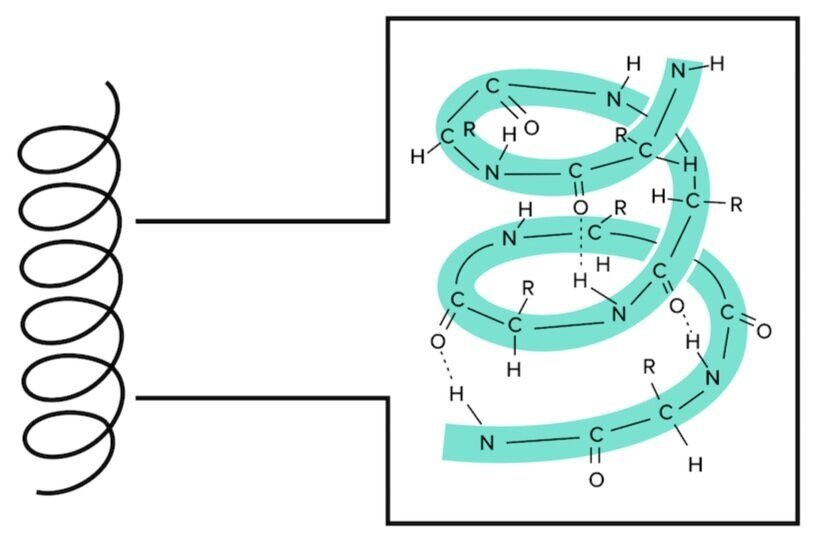
There are two main secondary structures: alpha helices and beta sheets. The alpha helix is stable because of the many hydrogen bonds that are formed when the backbone is arranged in this way. Notice that the R groups do not contribute to the hydrogen bonding forming the alpha helix.
Figure: An alpha helix is an extremely stable secondary structure.
Alpha helices serve a lot of different functions in different proteins. Many transmembrane proteins use alpha helices that span the entire membrane to transport ions from outside to inside the protein. You can find more information about transmembrane proteins in our guide on Lipids and Membranes.
Beta sheets are also formed through hydrogen bond interactions. However, instead of a helix arrangement, different regions on the amino acid string line up in rows.
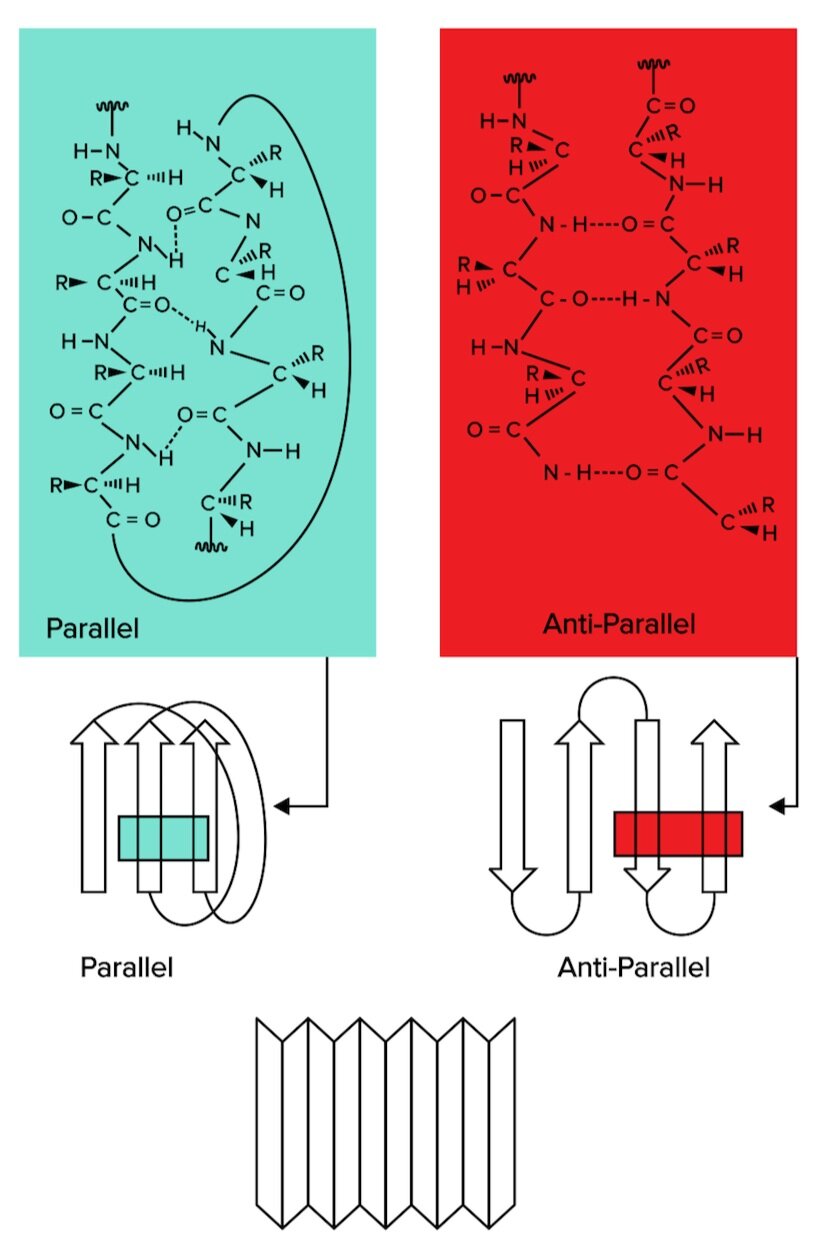
Figure: Beta sheets, another form of secondary structure, can be classified as parallel or antiparallel.
There are two types of beta sheets: parallel and antiparallel sheets. In parallel sheets, both strands of the polypeptide chain run in the same direction. In antiparallel sheets, the adjacent strands are running in the opposite directions (one is going from N to C terminus, while the other runs from C to N terminus).
Tertiary structure refers to structure that arises from interactions between the side chains of different amino acids. Positively charged side chains (histidine, lysine, and arginine) can interact with negatively charged side chains (aspartate, glutamate). Polar side chains will be attracted to other polar side chains. Nonpolar side chains will be attracted to other nonpolar side chains. Finally, two cysteine residues can covalently bond to each other, forming a disulfide bond.
Quaternary structure refers to interactions between two different protein subunits that make up a protein with more than one subunit. Many proteins have more than one subunit. Many proteins have more than ne subunit, or polypeptide strands. For instance, there are four subunits required to form hemoglobin. These interactions can be between backbone or between side chains. You will often find covalent disulfide bonds formed between two cysteine residues contributing to quaternary structure.
These bonds are formed after the protein has been completely translated from RNA. Such modifications are referred to as post-translational modifications and can include changes such as:
phosphorylation at certain amino acid residues (such as serine, threonine, and tyrosine)
formation of disulfide bonds between cysteine residues
glycosylation, or the linkage of short carbohydrate polymers to the protein
ubiquitination, or the attaching of a protein known as ubiquitin that marks the protein as a target for degradation
b) Specialized amino acids
Glycine
Recall that glycine possesses a single hydrogen as its side chain. Due to the relatively small size of this side chain, glycine is the least sterically hindered amino acid. As a result, it can rotate and move more easily. It is often found in areas of the protein which need a high amount of flexibility and rotation.
Cysteine
Recall that cysteine has a thiol group (S-H) at the end of the side chain. This gives cysteine the ability to bind to other cysteines through the formation of a disulfide bond (S-S). These disulfide bonds are important to both the tertiary and quaternary structures of many proteins. In particular, many proteins with multiple subunits form bonds between subunits using disulfide bonds.
Proline
In contrast to glycine, proline is the MOST sterically strained amino acid. Note that the side chain of proline is bound to the amino group of the amino acid. This creates a five-membered ring that does not have a lot of rotational ability and often creates a kink in the protein strand. This amino acid is often found in regions of the protein that should be immobile or when a bend in the structure is needed.
c) Stability and interactions
The multiple levels of structure within a protein are essential for the protein to function properly, particularly in the aqueous environment of the human body. Within this environment, the hydrophobic effect is an important force to consider.
The hydrophobic effect is a consequence of nonpolar and polar interactions. In an aqueous environment, hydrophobic residues will be attracted to each other, while they will also be repelled by the polar aqueous environment (water and ions in the water). For this reason, hydrophobic amino acids will be found in the interior of the protein rather than the outside of the protein.
This can also be explained from a thermodynamic standpoint. The hydrophobic effect also has to do with entropy. Most simply, entropy is a measure of the disorder of a system. Systems always increase in entropy, resulting in more disorder with time. If a protein has hydrophobic residues on the outside of the protein, then surrounding water molecules will have fewer residues to bind to on the surface of the protein.
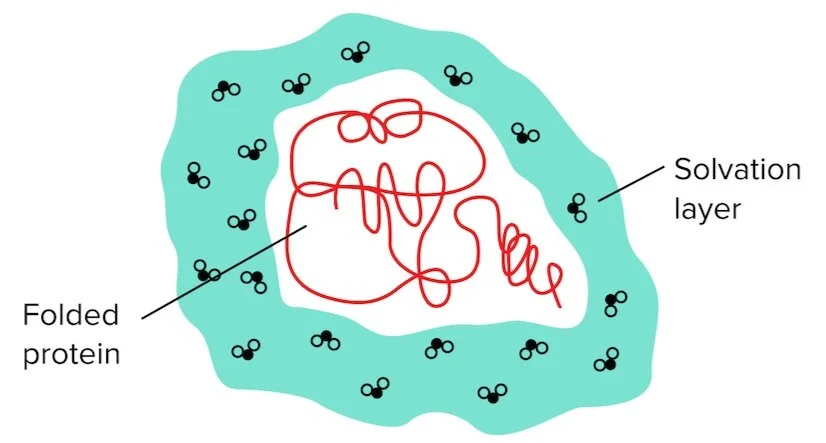
Figure: Water molecules surrounding the protein are known as the solvation/hydration layer.
When the outer protein residues are polar, the water molecules in the solvation layer can easily form hydrogen bonds in different rearrangements—water molecules in the solvation layer can hydrogen bond with polar residues and the other water molecules surrounding it.
However, if the outer layer of the protein is composed of mainly hydrophobic residues, the number of hydrogen bonds that the solvation layer can form becomes extremely limited. This requires the water molecules to be oriented more precisely/rigidly, resulting in an unfavorable reduction in entropy.
d) Protein folding
The solvation environment of a polypeptide chain directly influences the way a protein folds. Protein folding refers to the process through which a protein is organized (or folded) into its proper secondary and tertiary structures. Denaturing refers to the process through which a protein is unfolded or loses its proper 3D structure.
Protein folding can be disrupted by several environmental conditions, including:
High or low pH
High salt concentration (including the presence of molecules like urea)
High temperature
These same environmental changes can result in protein denaturation. As protein form is highly related to function, protein denaturation most often results in loss of function.
Denaturing a protein disrupts secondary, tertiary, and quaternary structures. Note that protein denaturation does not disrupt protein structure. In some proteins, it may be possible for a protein to refold back to its native state and regain its proper secondary, tertiary, and quaternary structure.
However, many proteins cannot refold to the native state once unfolded. Due to the high numbers of possible conformations that the polypeptide sequence can assume, folding a protein can be a tricky business. In biological systems, proteins called chaperones assist in folding denatured proteins back into their native state.
----
Part 5: High-yield terms
Amino group: a functional group composed of NH3+
Carboxylic acid: a functional group composed of COOH
Zwitterions: molecules that contain both positive and negative charges on the same molecule
Alpha carbon: central carbon atom of an amino acid, bonded to the amino group, carboxyl group, and R group
Peptide bond: bond between each amino acid in a protein; catalyzed by the ribosome; forms primary structure of a protein
Amide bond: another term for peptide bond; has partial double bond character which limits rotation of the constituent groups
Primary structure: the string of amino acids connected by peptide bonds, and is defined solely by the identity of amino acids within it
Secondary structure: formed through the hydrogen bonding interactions between atoms forming the backbone of the protein chain; includes alpha helices and beta sheets
Tertiary structure: structure that arises from interactions between the side chains of different amino acids
Quaternary structure: interactions between different protein subunits that make up a protein with more than one subunit
Hydrophobic effect: a consequence of nonpolar and polar interactions; in an aqueous environment, hydrophobic residues will be attracted to each other, while they will also be repelled by the polar aqueous environment
Protein folding: the process through which a protein is organized (or folded) into its proper secondary and tertiary structures
Denaturing: the process through which a protein is unfolded or loses its proper 3D structure
Chaperones: helper proteins that assist in folding denatured proteins back into their native state
----
Part 6: Passage-based questions and answers
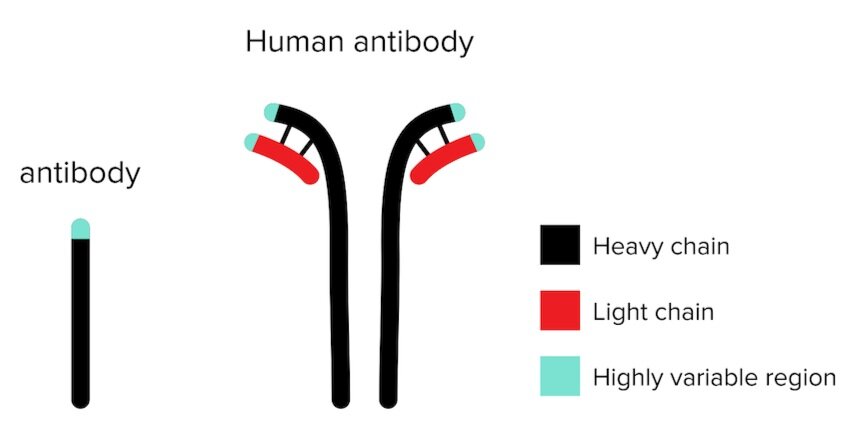
Figure 1: Mouse antibody and human antibodies.
Researchers inject mice with an antigen to induce the production of specific antibodies. Upon confirmation of antibody production, antibodies are isolated from blood samples taken from the mice and tested to determine if they are specific to the antigen using an enzyme-linked immunosorbent assay. Researchers also conjugate drugs to antibodies to produce drug conjugates called nanobodies. The nanobodies localize to antigens and often have a cleavable linker between the nanobody and drug, with an intention to localize nanobodies to the peripheral membrane protein. Once the nanobody localizes, it can enter the cell through endocytosis and then can be cleaved in a peroxisome or lysosome. Upon cleavage, the drug payload is released. Toxic drugs can be localized to cancer cells without causing undue damage to neighboring cells.
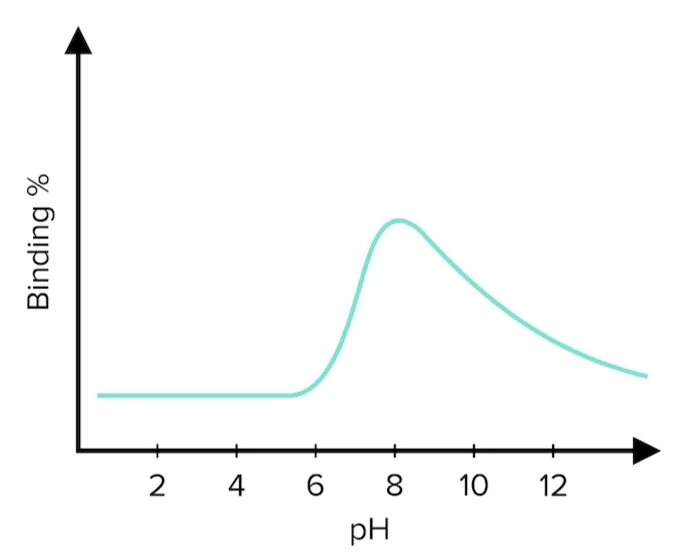
In a follow-up experiment, a nanobody identified as N96 successfully binds to HER2: an epidermal growth factor highly expressed in some breast cancers. Scientists have assayed the pH dependency of the N96’s affinity for HER2.
Figure 2: pH dependency of nanobody binding.
Question 1: Which of the following is likely to be found in human antibodies but not mouse nanobodies?
A) An antigen-specific variable region
B) Hydrophobic residues
C) Quaternary structure
D) Hydrogen bonding
Question 2: Researchers hope to identify the antigen-binding region of the nanobody. Based on the information provided in Figure 2, which of the following amino acids is most likely in this region?
A) Cys (pKa2 = 8)
B) Arg (pKa = 12)
C) Lys (pKa = 10)
D) Asp (pKa=3)
Question 3: Which of these residues is most likely to be found on the outside of N96?
A) Threonine
B) Phenylalanine
C) Leucine
D) Valine
Question 4: During drug delivery, the nanobody linker is cleaved by an intracellular protease that recognizes the pattern X-X-G-N-X-X, where X is any nonpolar amino acid residue. Which of the following amino acids should not be included in the linker?
A) Isoleucine
B) Valine
C) Glutamine
D) Phenylalanine
Answers to passage-based questions
Answer choice C is correct. Figure 1 provides a comparison of the structural differences between mouse and human antibodies. Mouse antibodies are composed of a single protein domain produced from a single mRNA transcript. These single subunits are functional on their own, without forming bonds to multiple subunits or creating a quaternary structure (choice C is correct). Both antibodies and nanobodies have antigen-specific variable regions (choice A is incorrect). All proteins are likely to have some amount of hydrophobic residues (choice B is incorrect). Hydrogen bonding is required to form secondary and tertiary structures, and thus provide protein function (choice D is incorrect).
Answer choice A is correct. According to Figure 2, nanobody binding peaks at pH=8. Its affinity gradually decreases from pH 7-12 and increases very quickly from pH 6-8. The pKa2 of cysteine is approximately 8, and this is the residue that indicates the highest binding affinity at pH=8 (choice A is correct). Both arginine and lysine are basic residues and have higher pKa values (choices B and C are incorrect). Aspartate, an acidic residue, would exhibit little to no binding affinity at its pKa (choice D is incorrect).
Answer choice A is correct. Threonine is a polar residue containing a hydroxyl (-OH) group (choice A is correct). Phenylalanine, leucine, and valine side chains contain nonpolar residues more frequently found folded on the interior of a protein (choices B, C, and D are incorrect).
Answer choice C is correct. The linker can include any nonpolar amino acids at the X positions. Isoleucine, valine, and phenylalanine are nonpolar residues that meet these criteria (choices A, B, and D are incorrect). Glutamine is a polar amino acid, so it cannot occupy the X positions (choice C is correct).
Want more MCAT practice questions? Check out our proprietary MCAT Question Bank for 4000+ sample questions and eight practice tests covering every MCAT category.
Gain instant access to 4,000+ representative MCAT questions across all four sections to identify your weaknesses, bolster your strengths, and maximize your score. Subscribe today to lock in the current investments, which will be increasing in the future for new subscribers.
----
Part 7: Standalone questions and answers
Question 1: Which of the following amino acids is naturally found in the R-configuration?
A) Tryptophan
B) Cysteine
C) Tyrosine
D) Methionine
Question 2: Why are hydrophobic residues often found on the interior of a protein?
A) They lower the entropy of the system
B) They are often less bulky than hydrophilic residues
C) Their van der Waals interactions are stronger than hydrogen bonds
D) The solvation layer is less ordered near hydrophilic residues
Question 3: Which of these is a form of primary structure interaction?
A) Disulfide bond
B) Peptide bond
C) Interactions between N-H and C=O of the protein backbone
D) Hydrophobic interactions
Question 4: Researchers hope to design a polypeptide inhibitor to a site with the repeated sequence A-Q-E-K-K. Which of these inhibitor sequences is most likely to succeed?
A) V-Q-R-A-A
B) E-Q-E-K-K
C) E-S-W-D-D
D) A-S-K-D-E
Question 5: Which of the following best describes the result of peptide bond hydrolysis?
A) The amino group of one amino acid attacks the carboxyl group of another amino acid
B) A water molecule is released into solution
C) A carboxylate group and an amine group are produced from an amide group
D) The protein has been denatured
Answers to standalone questions
Answer choice B is correct. R- and S-configuration refer to the chirality at the alpha carbon of the amino acid. Due to the priority rankings of all side chains, all 19 chiral amino acids are of the S-configuration except cysteine. This is due to the higher priority of the thiol side chain on the cysteine molecule, making it an R-configuration (choice B is correct). All other amino acids except glycine (which is achiral) are found in the S-configuration (choices A, C, and D are incorrect).
Answer choice D is correct. From a thermodynamic standpoint, the solvation layer is more ordered when nearby hydrophobic residues are nearby because there are fewer possible ways to create hydrogen bonds (choice D is correct). Hydrogen bonds are stronger than van der Waals interactions, and nonpolar residues cannot participate in hydrogen bonding (choice C is incorrect). Localization of hydrophobic residues has less to do with bulkiness than entropy; in fact, some hydrophilic chains such as aspartate and glutamate are quite bulky (choice B is incorrect). Hydrophobic residues lower the entropy of the system when on the exterior of the protein rather than the interior (choice A is incorrect).
Answer choice B is correct. Peptide bonds are formed between amino acids, and so comprise the primary structure of a protein (choice B is correct). Interactions between N-H and C=O groups of the protein backbone form alpha helices and beta sheets, forms of secondary structure (choice C is incorrect). Tertiary structure gives shape to globular proteins and arises from interactions between side chains of amino acids—including hydrophobic side chains (choice D is incorrect). Disulfide bonds between cysteine residues are found contributing to both tertiary structure and quaternary structure (choice A is incorrect).
Answer choice D is correct. To maximize attraction between the inhibitor and the site, the two sequences must exhibit similar attractions. The catalytic site, with a repeated sequence of alanine-glutamine-glutamate-lysine-lysine, has nonpolar-polar-acidic-basic-basic residues. As a result, the desired inhibitor should have: nonpolar-polar-basic-acidic-acidic residues to maximize hydrophobic interactions between nonpolar residues and ionic attractions between acidic and basic residues (choice D is correct). Placing alanine in the 4th and 5th positions would result in weak interactions with lysine (choice A is incorrect). Placing an acidic residue at position 1 would result in weak interactions with nonpolar alanine (choice B is incorrect). Tryptophan is not a charged residue, and so would not be paired well with glutamate (choice C is incorrect).
Answer choice C is correct. Peptide bonds are formed through nucleophilic attack (choice A is incorrect). Peptide bond formation also results in dehydration, or the release of one water molecule (choice B is incorrect). Denaturation occurs when secondary, tertiary, and quaternary structures are disrupted rather than primary structure (choice D is incorrect).